Making Tutorials¶
This project was supported by grants from the National Institute of Biomedical Imaging and Bioengineering (U24EB029012) from the National Institutes of Health.
Authors:
Jess Tate
Overview¶
This tutorial demonstrates how to use markdown to create new tutorials for UncertainSCI. It will walk through all the files needed and the basic structure needed expected for tutorials. Knowledge of Markdown, Github, Python, Sphinx, and Read the Docs will be useful. If you have questions, please ask.
Software requirements¶
UncertainSCI¶
To make a Tutorial for UncertainSCI, start with an up-to-date version of the code and documentation. Download the source code or clone the repository from github. We suggest creating a fork of the repository so that you can track your changes and create pull requests to the UncertainSCI repository. UncertainSCI requirements are found here
Dependencies and Development Tools¶
UncertainSCI uses Read the Docs and Sphinx to build and host tutorial documentation. This platform converts markdown files to html for web viewing using Sphinx, a Python Library. Testing the new documentation may require building the web pages locally for viewing. This will require installing Python, pip, Sphinx, Recommonmark, and other packages in the docs/requirements.txt file. More information can be found in the walkthorugh and on the Sphinx documentation.
Creating Your UncertainSCI Fork¶
With your own github account, go to the UncertainSCI Github page. Click the fork button on the upper right side of the page. It will ask you where to move the fork to, chose your own account. Once the repository is forked, clone it to your local machine with the following command.
$git clone https://github.com/[yourgithubaccount]/UncertainSCI.git
After the the code is cloned, navigate to the repository directory and add the upstream path to the original UncertainSCI repository.
$git remote add upstream https://github.com/SCIInstitute/UncertainSCI.git
You should be able to see both your and the original repositories when you use the command:
$git remote -v
The fork is good to go, but you will need to sync the fork occasionally to keep up with the changes in the main repository. To sync your fork, use the following commands:
$git fetch upstream
$git checkout master
$git merge upstream/master
You should sync and merge your fork before you start a new module and before you create a pull request.
It is a good practice to create a new branch in your fork for every module you will be adding. The command to create a new branch is:
$git checkout -b [branch_name]
Please see the Github help page for more information.
Files Needed for a New Tutorial¶
This chapter will describe the files need to create a Tutorial for UncertainSCI.
Overview of Files Needed for a Tutorial¶
To make a new tutorial, a markdown file is required for the content of the tutorial. Other files, such as images, may also be included. In addition to the new files for the tutorial, a link to the new tutorial should be added to the User Documents file.
Markdown File¶
The main file needed for a new tutorial is a markdown file. The file should have an file ending of .md and should be located in the UncertainSCI/docs/tutorials/ directory. There is a template file that can be used, or an existing tutorial like this one can be used.
If using math equations in a latex, a call to the mathjax server is required at the begining of the document:
<script type="text/javascript" async
src="https://cdn.mathjax.org/mathjax/latest/MathJax.js?config=TeX-AMS_CHTML">
</script>
Sphinx will build the menu and table of contents from the header names, and the document will be formated correctly if the proper markdown headings levels are used. For instance, the Document title should be the first content:
# Title
Then the chaper headings:
## Overview
## Chapter The First
## ETC.
An important part of the document is to acknowledge the authors and funding sources. Include text similar to this after the document title:
This project was supported by grants from the National Institute of Biomedical Imaging and Bioengineering (U24EB029012) from the National Institutes of Health.
Authors:
Smart people here
Chapters and sections will appear in the table of contents with the page titles. Once the file is added to the docs/tutorial/index.rst file
Added Figures¶
Most tutorials require a number of screenshots and other images. Figures and images should be added in a folder for each tutorial in the UncertainSCI/docs/user_docs/ folder. The figure folder should be named after the tutorial, for example, the images in this tutorial are in a folder called HowToTutorial_figures. Instructions on how to use the images and figures are found here
Additional Files¶
Additional files added to the tutorials folder should be minimized as much as possible. Example scripts should be located in the UncertainSCI/examples directory and example data will, generally, need separate hosting. However, if one or two files are needed, they may be added to the UncertainSCI/docs/tutorials/ with a consistent naming scheme. Bibtex file with a matching name should be added in UncertainSCI/docs/tutorials/, yet if multiple additional files are needed, they should placed in a new folder indicating the tutorial: UncertainSCI/docs/tutorials/[tutorial name]_files.
Linking to New Tutorial¶
For the new tutorial to be visible on the tutorials page, add the filename to the list in the UncertainSCI/docs/tutorials/index.rst file.
Testing Documentation¶
This chapter describes how to test the look and content of the new tutorial. Test the generated github-pages with either a local Sphinx build or using the online build on Read the Docs.
Testing Locally¶
Testing the documentation locally involves building documentation website on your local machine using Sphinx and Recommonmark. These instructions are adapted from Read the Docs’ help page.
Installing Shpinx¶
To install the Sphinx, make sure that a relatively recent version of Python 3. Installing sphinx and other dependencies is easiest with pip, just run
pip install -r requirements.txt
in the UncertainSCI/docs folder and the relavent python dependencies, including Sphinx, will be installed.
Alternatively, the dependencies listed in UncertainSCI/docs/requirements.txt file could be installed seperately.
Please ask if you have any questions.
Building Documentation¶
Once the relavent python packages are installed properly, such as with pip, building the documentation pages is as simple as running
make html
in the UncertainSCI/docs folder. A script will run and, assuming no errors, the html pages will be generated within the UncertainSCI/docs/_build/ folder. Any of the html files can be tested by opening it in a browser.
Testing Online¶
In addition to building and testing the documentation pages locally, they can also be tested online with Read the Docs. Any branch of a github repo can be built one the Read the Docs page. Online builds can only be triggered deirectly on the readthe docs by those who have access. However, every pull request to the UncertainSCI repo will trigger an online build that can be viewed by anyone.
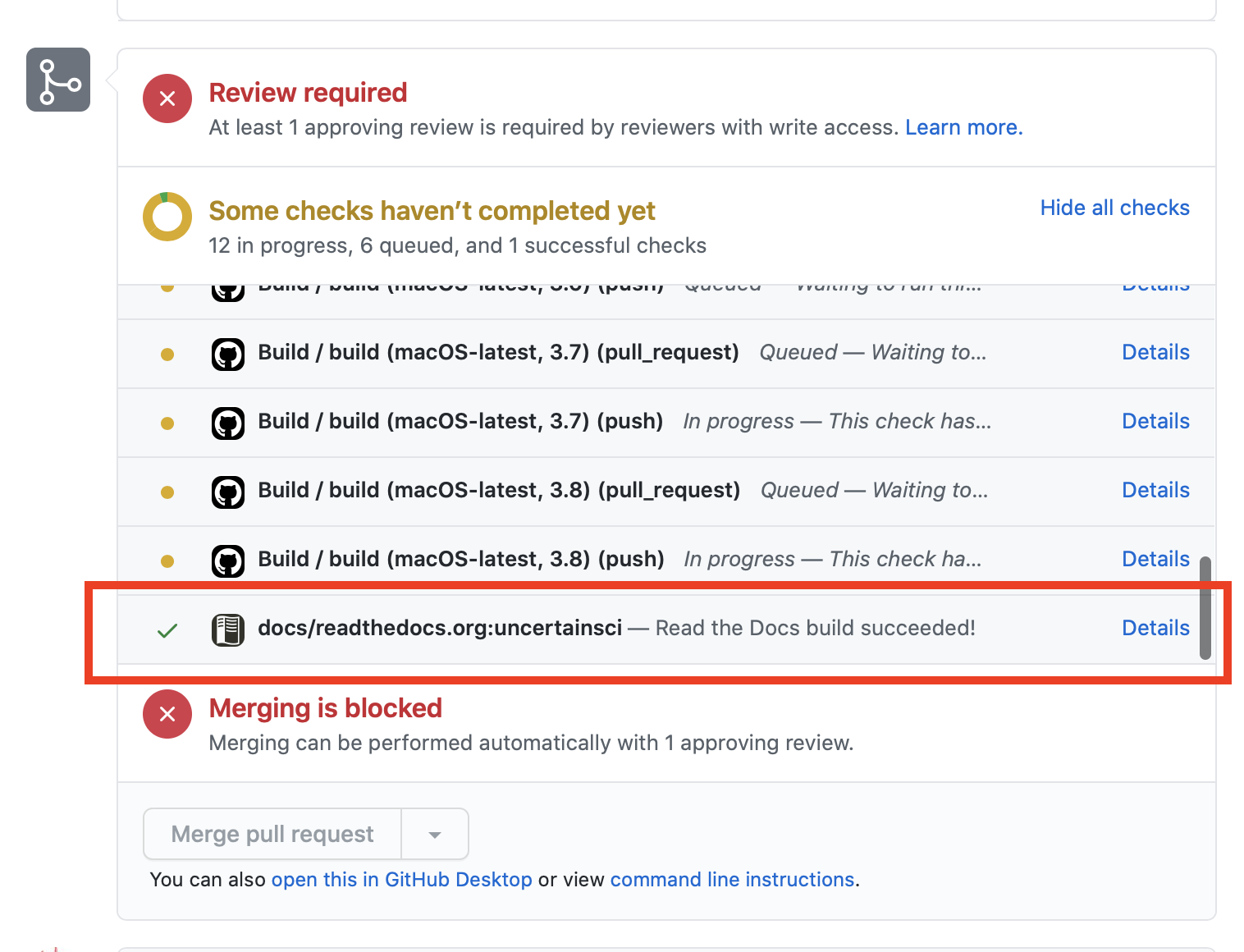 The documentation page can be viewd by clicking on the details link, shown in the image. Pull requests are required when submitting contributions to UncertainSCI.
The documentation page can be viewd by clicking on the details link, shown in the image. Pull requests are required when submitting contributions to UncertainSCI.
Please ask if you have any questions.
Adding Content¶
This chapter provides some examples of how to add some types of content that may be needed for a tutorial. For general Markdown info, see here and here.
Figures¶
Figures can be added fairly easily in Markdown, with a simple call to the location:

However, using a bit of html allows us to reference the figure easier:
<figure id="example">
<img src="../_static/UncertainSCI.png" alt="UncertainSCI example image">
<figcaption>Example for including an image in tutorial.</figcaption>
</figure>
And to reference (not working with Sphinx):
<a href="#example">reference the figure</a>
Math¶
Math equations can be used in Markdown using MathJax. Mathjax will convert LaTex format:
$$ \frac{\partial \rho}{\partial t} + \nabla \cdot \vec{j} = 0 \,. \label{eq:continuity} $$
$$ \frac{\partial \rho}{\partial t} + \nabla \cdot \vec{j} = 0 ,. \label{eq:continuity} $$ It can also use MathJax specific tags:
\\[ x = {-b \pm \sqrt{b^2-4ac} \over 2a} \\]
\[ x = {-b \pm \sqrt{b^2-4ac} \over 2a} \]
inline equations use the \\(\mathbf{p}\\) sytanx: \(\mathbf{p}\)
Citations¶
Since we are using Sphinx to build the documentation, we can use its citation manager, sphinxcontrib-bibtex. We will provide some exmples here, but for more information, refer to the sphinxcontrib-bibtex documentation.
Citations to include in the UncertainSCI docs can be included in the UncertainSCI/docs/references.bib file. For the keys, please use the convention: Initials of the contributor, colon, first three letters of the author (with apropriate capitalization), and the year. For example JDT:Bur2020. For multiple citations from the same author in the same year, lowercase letters can be appended to the key: JDT:Bur2020a.
After the reference has been added to UncertainSCI/docs/references.bib, the final step is to include the command in the appropriate place.
The sphinxcontrib-bibtex is built to run with rst in Sphinx. However, it can be used in markdown using the AutoStructify package. This will require using using an eval_rst block as follows:
```eval_rst
The whole paragraph will need to be in the eval_rst block :cite:p:`JDT:Bur2020`. For multiple references: :cite:p:`JDT:Bur2020,gupta1983`
```
The whole paragraph will need to be in the eval_rst block [2]. For multiple references: [2, 6]
add a bibliography section
```eval_rst
.. bibliography:: ../references.bib
```
Snippets¶
Basic markdown some snippet
def function():
return True
def function():
return True
Links¶
Including links in Markdown is simple, just use <> or [](). For example, an internal link for section Adding Content is :
[Adding Content](#adding-content)
When using internal links to sections, include the name of the section, all lower case and with - replacing spaces, and all special characters ommited. Linking to other pages in within the UncertainSCI documentation requires a relative path. Demos is:
[Demos](../tutorials/demos.html#demos)
Links to other websites can include the full URL. Using <> will show the URL, []() will hide it with other text.
<https://www.markdownguide.org>
[Markdown](https://www.markdownguide.org)
Tables¶
Tables can be used with normal markdown syntax with the sphinx-markdown-tables package
| Syntax | Description |
| ----------- | ----------- |
| Header | Title |
| Paragraph | Text |
| Syntax | Description |
|---|---|
| Header | Title |
| Paragraph | Text |
Header 1 |
Header 2 |
Header 3 |
|---|---|---|
body row 1 |
column 2 |
column 3 |
body row 2 |
Cells may span columns. |
|
Referencing Sphinx¶
To link the UncertainSCI API generated using Sphinx, Use this syntax: [text](../api_docs/pce.html#polynomial-chaos-expansions).
Content Guide¶
Try to be comprehensive, yet consise, and keep in mind the target audience for the tutorial. It is ok to write an advanced tutorial that builds on more basic knowledge, but please make this expectation clear and link to tutorials and materials that will help the reader develop the required understanding. Include code snippets, example scripts, screenshots, and videos as appropriate. Please use existing tutorials as try to match the style, flow, and level of detail they provide.
Supplemental Materials¶
Some things to consider including with the tutorial.
Example Scripts¶
Example scripts should be located in the UncertainSCI/demos/ directory. Consider using one of the existing demos as a template and try to follow the coding standards outlined in the contribution guide.
Movies¶
Movies should be stored in a serperate place. We host most of ours on youtube or vimeo.
Youtube¶
Get the imbed link from the youtube video. This can be found by pressing the share button.
Vimeo¶
Get the imbed link from the vimeo video. This can be found by pressing the share button. There are more options if for some users who own the video. More info here.
With the embed link, just include the html into the markdown file.
<iframe title="vimeo-player" src="https://player.vimeo.com/video/279319572" width="640" height="360" frameborder="0" allowfullscreen></iframe>
Datasets¶
Datasets should be located serperately, unless the size is small. Please ask if you have any questions.
Bibliography¶
Akil Narayan, Zexin Liu, Jake Bergquist, Chantel Charlebois, Sumientra Rampersad, Lindsay Rupp, Dana Brooks, Dan White, Jess Tate, and Rob S MacLeod. UncertainSCI: uncertainty quantification for computational models in biomedicine and bioengineering. Available at SSRN 4049696, 2022.
Kyle M. Burk, Akil Narayan, and Joseph A. Orr. Efficient sampling for polynomial chaos-based uncertainty quantification and sensitivity analysis using weighted approximate fekete points. International Journal for Numerical Methods in Biomedical Engineering, 36(11):e3395, 2020. doi:https://doi.org/10.1002/cnm.3395.
Akil Narayan. Computation of induced orthogonal polynomial distributions. Electronic Transactions on Numerical Analysis, 50:71–97, 2018. arXiv:1704.08465 [math]. URL: https://epub.oeaw.ac.at?arp=0x003a184e, doi:10.1553/etna_vol50s71.
L. Guo, A. Narayan, L. Yan, and T. Zhou. Weighted Approximate Fekete Points: Sampling for Least-Squares Polynomial Approximation. SIAM Journal on Scientific Computing, 40(1):A366–A387, 2018. arXiv:1708.01296 [math.NA]. URL: http://epubs.siam.org/doi/abs/10.1137/17M1140960, doi:10.1137/17M1140960.
Albert Cohen and Giovanni Migliorati. Optimal weighted least-squares methods. SMAI Journal of Computational Mathematics, 3:181–203, 2017. arxiv:1608.00512 [math.NA]. doi:10.5802/smai-jcm.24.
S.K. Gupta and W.V. Harper. Sensitivity/Uncertainty Analysis of a Borehole Scenario Comparing Latin Hypercube Sampling and Deterministic Sensitivity Approaches. Technical Report BMI/ONWI-516, Battelle Memorial Institute, Office of Nuclear Waste Isolation, 1983.
Jake Bergquist, Brian Zenger, Lindsay Rupp, Akil Narayan, Jess Tate, and Rob MacLeod. Uncertainty quantification in simulations of myocardial ischemia. In Computing in Cardiology, volume 48. September 2021.
Lindsay C Rupp, Zexin Liu, Jake A Bergquist, Sumientra Rampersad, Dan White, Jess D Tate, Dana H. Brooks, Akil Narayan, and Rob S. MacLeod. Using uncertainSCI to quantify uncertainty in cardiac simulations. In Computing in Cardiology, volume 47. September 2020.
Lindsay C Rupp, Jake A Bergquist, Brian Zenger, Karli Gillette, Akil Narayan, Jess Tate, Gernot Plank, and Rob S. MacLeod. The role of myocardial fiber direction in epicardial activation patterns via uncertainty quantification. In Computing in Cardiology, volume 48. September 2021.
Jess D. Tate, Wilson W. Good, Nejib Zemzemi, Machteld Boonstra, Peter van Dam, Dana H. Brooks, Akil Narayan, and Rob S. MacLeod. Uncertainty quantification of the effects of segmentation variability in ECGI. In Functional Imaging and Modeling of the Heart, pages 515–522. Springer-Cham, Palo Alto, USA, 2021. doi:https://doi.org/10.1007/978-3-030-78710-3_49.
Jess Tate, Sumientra Rampersad, Chantel Charlebois, Zexin Liu, Jake Bergquist, Dan White, Lindsay Rupp, Dana Brooks, Akil Narayan, and Rob MacLeod. Uncertainty quantification in brain stimulation using uncertainSCI. Brain Stimulation: Basic, Translational, and Clinical Research in Neuromodulation, 14(6):1659–1660, January 2021. URL: https://doi.org/10.1016/j.brs.2021.10.226, doi:10.1016/j.brs.2021.10.226.
Jess D Tate, Nejib Zemzemi, Shireen Elhabian, Beáta Ondru\u sová, Machteld Boonstra, Peter van Dam, Akil Narayan, Dana H Brooks, and Rob S MacLeod. Segmentation uncertainty quantification in cardiac propagation models. In 2022 Computing in Cardiology (CinC), volume 498, 1–4. 2022. doi:10.22489/CinC.2022.419.